SARS-CoV-2 Diagnostics and Addressing The Needs of The Pandemic
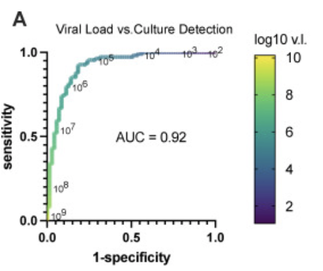 Figure of Receiver Operator Characteristics comparing Viral Load and Culture. A viral load of > 10E5 suggests infectivity
Figure of Receiver Operator Characteristics comparing Viral Load and Culture. A viral load of > 10E5 suggests infectivity
Swabs and Supply Chain. There were insufficient swabs of the appropriate type to sustain current COVID19 testing efforts. See collaborative efforts to produce, test, and evaluate nasopharyngeal swabs made through use of 3D-printing technology. Colleague Ramy Arnaout spearheading these efforts assisted by Kirby Lab members. Pictures of the innovative swab designs examined may be found on GitHub. We addressed the shortage of viral transport medium by setting up a multi-shift assembly line for viral transport medium and collection kit production.
Sample types. Several additional studies provided comparison among various samples types (nasopharyngeal swabs, nasal swabs, and saliva). Our general find was the nasal swabs performed just as well as nasopharyngeal swabs for patients with viral loads > 1000 genome copies/mL. As this is well below the infectious level for patients, these less invasive samples types for practical purposes are equivalent.
Disinfection of N-95 Masks. We also established a microwave-based steam method using commonly available household items to disinfect N95 masks in 3 minutes, using the MS2 RNA phage as a SARS-CoV-2 viral surrogate, as described in mBio. As of August 2021 this was this most read manuscript in this journal based on online metrics.
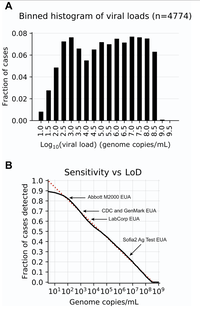
Evaluation and comparison of PCR, Antigen Test, and Culture Diagnostics. We have most recently been collaborating with Phyllis Kanki's laboratory at the Harvard School of Public Health to compare the performance of RT-qPCR and antigen tests with viral culture for SARS-CoV-2 detection. We found, as published in Clinical Microbiology and Infection, that antigen tests are excellent at identifying those who are culture positive and therefore infectious, and outstanding at identifying the most infectious individuals. We are grateful for support form the Massachusetts Life Science Center, Accelerating Coronovirus Testing Solutions grant for this work.
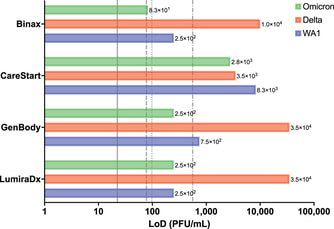
Our studies further support use of antigen tests in our ongoing response to the COVID-19 pandemic. In collaboration with graduate student, Sydney Stanley, and the Kanki Laboratory we also determined that commonly used antigen tests performed just as well against the Omicron variant as the original Washington strain. Importantly, our limit of detection studies were performed with live virus which likely allows a more real-life comparison of assay performance.
The Rationale for Reporting out SARS-CoV-2 Viral Loads, NOT Ct values. We believe strongly that cycle thresholds are non-ideal to report out as viral loads may vary over 1000-fold for any given Ct value depending on the commercial SARS-CoV-2 RT-PCR method. This has been borne out in a multi-institutional collaborative study. Furthermore, assays should be calibrated against a universal standard and reported out as a viral load. The absolute and comparative limit of detection of methods does matter.
Our cumulative work led to awarding of one of two in the national Reagan-Udall awards with Ramy Arnaout as lead PI. The goal of this award was to use real-world data to inform the regulator path way for SARS-CoV-2 antigen and PCR EUA assays.
Sample types. Several additional studies provided comparison among various samples types (nasopharyngeal swabs, nasal swabs, and saliva). Our general find was the nasal swabs performed just as well as nasopharyngeal swabs for patients with viral loads > 1000 genome copies/mL. As this is well below the infectious level for patients, these less invasive samples types for practical purposes are equivalent.
Disinfection of N-95 Masks. We also established a microwave-based steam method using commonly available household items to disinfect N95 masks in 3 minutes, using the MS2 RNA phage as a SARS-CoV-2 viral surrogate, as described in mBio. As of August 2021 this was this most read manuscript in this journal based on online metrics.
Evaluation and comparison of PCR, Antigen Test, and Culture Diagnostics. We have most recently been collaborating with Phyllis Kanki's laboratory at the Harvard School of Public Health to compare the performance of RT-qPCR and antigen tests with viral culture for SARS-CoV-2 detection. We found, as published in Clinical Microbiology and Infection, that antigen tests are excellent at identifying those who are culture positive and therefore infectious, and outstanding at identifying the most infectious individuals. We are grateful for support form the Massachusetts Life Science Center, Accelerating Coronovirus Testing Solutions grant for this work.
Our studies further support use of antigen tests in our ongoing response to the COVID-19 pandemic. In collaboration with graduate student, Sydney Stanley, and the Kanki Laboratory we also determined that commonly used antigen tests performed just as well against the Omicron variant as the original Washington strain. Importantly, our limit of detection studies were performed with live virus which likely allows a more real-life comparison of assay performance.
The Rationale for Reporting out SARS-CoV-2 Viral Loads, NOT Ct values. We believe strongly that cycle thresholds are non-ideal to report out as viral loads may vary over 1000-fold for any given Ct value depending on the commercial SARS-CoV-2 RT-PCR method. This has been borne out in a multi-institutional collaborative study. Furthermore, assays should be calibrated against a universal standard and reported out as a viral load. The absolute and comparative limit of detection of methods does matter.
Our cumulative work led to awarding of one of two in the national Reagan-Udall awards with Ramy Arnaout as lead PI. The goal of this award was to use real-world data to inform the regulator path way for SARS-CoV-2 antigen and PCR EUA assays.