Emerging antimicrobial resistance is threatening our ability to treat bacterial infection. The Infectious Disease Society of America indicates that in the next decade we will lose our ability to treat the most common causes of bacterial infection, the so called ESKAPE pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumanii, Pseudomonas aureugosa, and Enterobacter sp. The US Centers for Disease Control and Prevention additionally cite carbapenem-resistant Enterobacteriaceae (also know as CRE) among the most serious antibiotic resistance threats. New antimicrobials to treat these multi-drug resistant pathogens are urgently needed.
Therefore, the Kirby Laboratory, funded by grants form the National Institutes of Health, is taking a number of approaches to identify and develop next generation antimicrobials. Our approaches build on an understanding of how bacterial pathogens causes disease and evade therapy by current antibiotics.
Therefore, the Kirby Laboratory, funded by grants form the National Institutes of Health, is taking a number of approaches to identify and develop next generation antimicrobials. Our approaches build on an understanding of how bacterial pathogens causes disease and evade therapy by current antibiotics.
A sampling of current antimicrobial projects:
(1) Attacking the biology of Gram negative infection.
Type IV secretion systems (T4SS) are syringe like mechanisms used to inject virulence factors into hosts cells. These systems are absolutely required for virulence in Gram negative pathogens such as Legionella pneumophila, Brucella, Bartonella, Coxiella burnetii, Rickettsia, Anaplasma, Ehrlichia, and Helicobacter pylori. Therefore, we predict that small molecule inhibitors of T4SS should render such organisms avirulent. In doing so, they will serve as a new type of therapeutic agent -- that blocks virulence rather than directly killing the pathogen directly. They will also serve as new scientific tools to explore the biology of these important bacterial virulence strategies. To date, we have used Legionella pneumophila as a model system to identify and establish proof of principle for type IV secretion system-based therapeutics. Using a high throughput screening approach, several lead candidates and scaffolds have been identified. We are exploring the relationship between activity and structure of compounds to define important structure motifs, so called structure-activity relationship (SAR) studies. We are also characterizing activity of compounds in several infectious model systems.
(2) Screening for inhibitors that act on the pathogen, pathogen-host interface, and/or host cells to limit intracellular bacterial replication.
|
We recently described a novel high throughput screening technology (highlighted on the cover of the May issue of Assay Development and Technology) to measure contemporaneously and in real-time effects on both intracellular growth of Legionella and host cell viability. This methodology has allowed us to rapidly screen 200,000+ compounds for intracellular growth inhibition, while at the same time weeding out compounds that are toxic to eukaryotic cells. In this way, our screening hits are enriched for those with therapeutic antimicrobial potential.
|
(3) Combating multi-drug resistance
 Log-fold reduction in intracellular growth of Legionella in high throughput screening assay
Log-fold reduction in intracellular growth of Legionella in high throughput screening assay
We are developing high throughput screening technologies to identify small molecules that restore antimicrobial susceptibility in multi-drug resistance strains, specifically carbapenem-resistant Enterobacteriaceae (CRE), a CDC-designated urgent antimicrobial resistance threat. These include multidrug-resistant Escherichia coli, Klebsiella, and Enterobacter species. CRE are defined, as their name implies, by their resistance to carbapenems, a broad spectrum antimicrobial therapy, often the agent of last resort in multidrug-resistant Gram-negative infections. Carbapanems are beta-lactam antibiotics. Resistance is usually related to the activity of carbapenemase enzymes encoded on large multidrug resistance plasmids. Resistance to carbapenems generally confers resistance to all other beta-lactams including penicillins and cephalosporins. During the past several years we have seen significant emergence of CRE in the United States and throughout the world. Often CRE remain susceptible to only toxic antimicrobials such as colistin or aminoglycosides (kidney failure, hearing loss) and sometimes are resistant to all clinically available antimicrobials. Restoration of susceptibility to non-toxic agents such as carbapenems would therefore provide considerable clinical benefit.
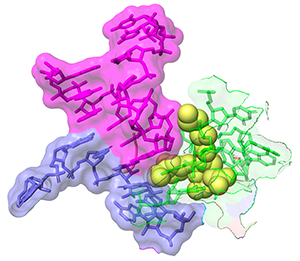
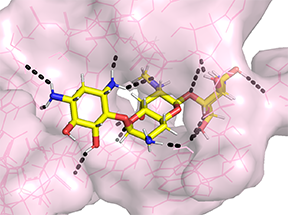 Apramycin bound to 16S rRNA helix 44 from PDB 7PJS
Apramycin bound to 16S rRNA helix 44 from PDB 7PJS
As an example of our screening efforts in this area, KP Smith, a postdoctoral fellow in the Kirby laboratory, recently published a validation of a high throughput screening strategy to identify small molecules that either directly inhibit CRE or potentiate the activity of meropenem against CRE. Interestingly, several existing human drugs (AZT, spectinomycin, apramycin) showed excellent activity, with fairly broad activity spectrum against a large panel of highly resistant CRE strains tested. In fact, we found that overt resistance to apramycin was virtually absent in not only CRE, but also multidrug-resistant, extensively drug-resistant, and pandrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Furthermore, we found that apramycin has potent activity against tissue infection with highly resistant Acinetobacter baumannii. Therefore, these and related agents warrant further study. They potentially could be repurposed for treatment of CRE and other multidrug-resistant pathogens or serve as the starting points for medicinal chemistry to seek derivatives with more favorable activity profiles. We are collaborating with the medicinal chemistry groups of Roman Manetsch and George O'Doherty at Northeastern University to further explore compound "scaffolds" identified through our high throughput screening efforts with promising activity against multidrug-resistant CRE, Acinetobacter baumannii, and :Pseudomonas aeruginosa.
Separately, we participate in a multi-institutional collaboration spearheaded by the Broad Institute to sequence the genomes of CRE and are collaborating with Ashlee Earl's group at the Broad Institute to define mechanisms of resistance.
Separately, we participate in a multi-institutional collaboration spearheaded by the Broad Institute to sequence the genomes of CRE and are collaborating with Ashlee Earl's group at the Broad Institute to define mechanisms of resistance.
(4) Developing natural products as next generation Gram-negative antimicrobials
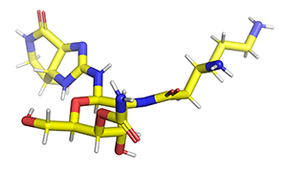 streptothricin F
streptothricin F
Micro-organisms produce antibiotics to combat other micro-organisms and gain turf. We have borrowed these antibiotics from this internecine arm's race and repurposed them for combating human and veterinary infections. Unfortunately, micro-organisms have already developed defenses against such antimicrobials, often in the form of antibiotic-modifying enzymes that can be horizontally transferred to pathogens we are trying to treat. This has led to emerging antimicrobial resistance to the extent that an increasing number of Gram-negative infections may be effectively untreatable with available antimicrobial agents. We have several efforts whose goal is to either to develop ignored antibiotic scaffolds with unique mechanisms of action, or through chemical modification of an exisitng antibiotic class render it immune to existing resistance mechanisms. This research combines the efforts of four collaborating laboratories (Kirby Lab-microbiology; Manetsch and O'Doherty laboratories, Northeastern University, medicinal chemistry; Yu Lab, Case Western Reserve University, structural biology).
For example, in 1492, Waksman and colleagues, identified a compound class known as streptothricins. Initially, streptothricins generated intense interest, as they were the first antibiotic class identified with potent activity against Gram-negative pathogens. However, limited testing in humans was associated with reversible kidney toxicity. Streptomycin was soon thereafter discovered, relegating streptothricins pretty much to the history books. We took another look at streptothricins and found outstanding activity against carbapenem-resistant Enterobacterales and Acinetobacter baumannii, including pan-drug resistant strains. In particular, a component of the streptothricin natural product discovered by Waksman, what we now know is a mixture of many structurally related streptothricin molecules, was found to have compelling biological activity both in vitro and in vivo. Moreover, we found that its target is unique and different from other known protein translation inhibitors binding to helix 34 of the 16S rRNA of the small ribosomal subunit. More details are described in our study of Plos Biology. Its binding immediately next to the site of the A-site tRNA provides a plausible explanation for the potent miscoding and resulting rapid bactericidal activity of this natural product.
We are collaborating with the labs of Roman Manetsch and Ed Yu to investigate the structural space around this natural product scaffold and the structural biology of its interactions with the ribosome. Roman's laboratory has developed a total de novo synthesis for streptothricin which supports the exploration of structural activity relationships of the streptothricin scaffold with the goal of finding compounds with enhanced activity that avoids existing resistance associated with acetylation of the beta-amine on the beta-lysine moiety in the molecule by nourseothricin acetyl transferase enzymes found in some strains.
For example, in 1492, Waksman and colleagues, identified a compound class known as streptothricins. Initially, streptothricins generated intense interest, as they were the first antibiotic class identified with potent activity against Gram-negative pathogens. However, limited testing in humans was associated with reversible kidney toxicity. Streptomycin was soon thereafter discovered, relegating streptothricins pretty much to the history books. We took another look at streptothricins and found outstanding activity against carbapenem-resistant Enterobacterales and Acinetobacter baumannii, including pan-drug resistant strains. In particular, a component of the streptothricin natural product discovered by Waksman, what we now know is a mixture of many structurally related streptothricin molecules, was found to have compelling biological activity both in vitro and in vivo. Moreover, we found that its target is unique and different from other known protein translation inhibitors binding to helix 34 of the 16S rRNA of the small ribosomal subunit. More details are described in our study of Plos Biology. Its binding immediately next to the site of the A-site tRNA provides a plausible explanation for the potent miscoding and resulting rapid bactericidal activity of this natural product.
We are collaborating with the labs of Roman Manetsch and Ed Yu to investigate the structural space around this natural product scaffold and the structural biology of its interactions with the ribosome. Roman's laboratory has developed a total de novo synthesis for streptothricin which supports the exploration of structural activity relationships of the streptothricin scaffold with the goal of finding compounds with enhanced activity that avoids existing resistance associated with acetylation of the beta-amine on the beta-lysine moiety in the molecule by nourseothricin acetyl transferase enzymes found in some strains.
(5) High throughput synergy testing
Analysis of antimicrobial susceptibility of intracellular pathogens such as Legionella pneumophila has been problematic. Traditional broth dilution susceptibility testing does not fully address access of antimicrobials to the intracellular replicative niche. Therefore, initial suggestion of therapeutic efficacy of beta-lactam antibiotics proved clinically incorrect. Assessment of effects on intracellular replication is technically complex. However, using high throughput screening technology developed in our laboratory, we were recently able to analyze the effects of ~250 known antibiotics against intracellular growth of Legionella pneumophila. Results correlated with known therapeutic efficacy against Legionnaires' disease. For example, later generation macrolides, but not erythromycin, were potent, as were almost all quinolones. Interestingly, previously under-investigated antimicrobials in Legionnaires' disease such as minocycline and florfenicol showed excellent potency. Furthermore, taking advantage of digital dispensing robotics, we were able to address two and three dimensional synergy for the most potent antimicrobials against intracellular growth.
|
What is synergy? Synergy is the ability of two or more antimicrobials when you used together to have much greater effect than the sum of their separate effects. Or in other words: A + B (observed) >> A+B (expected).
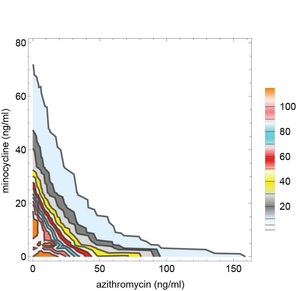
Sometimes synergy is graphically represented by plotting the permutations of antibiotic concentrations that results in inhibition of bacterial growth. These plots are known as isobolograms. Using such analysis, we found that pairwise combinations of azithromycin, minocycline, and rifampin showed strong synergy. See example on the left showing pairwise synergy between minocycline and azithroymycin. In isobologram plots, a straight line connecting the minimal inhibitory concentration (MIC) of each drug indicates indifference, while a concave isobologram curve, such as the one shown, indicates synergy. In this plot, the right most contour line connects points of 99% inhibition relative to intracellular growth of untreated control cultures. Additional contour lines connecting antibiotic concentrations the lead to partial levels of intracellular growth inhibition are also shown. (Isobologram plots to the left and below were drawn with Mathematica. See our recent publication for additional details.) |
|
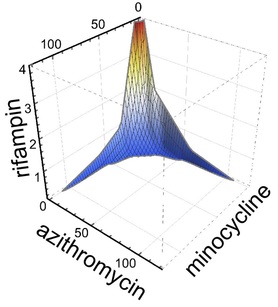
Intriguingly, when used in triple combination, azithromycin, minocyline, and rifampin showed even more potent three dimensional synergy. Specifically, when used in triple combination, only one tenth the concentration of each antimicrobial was required to inhibit intracellular growth compared to when each antimicrobial was tested separately. See graph to the right showing an isobologram surface, connecting points of 99% intracellular growth inhibition, demonstrating a high degree of surface concavity. It is intriguing to consider therapeutic implications and application of this data, especially in disease processes such as lung consolidation where penetrance of antimicrobials may be less than ideal. Just as importantly we found that combinations that might be used in severe community acquired pneumonia such as ceftriaxone plus azithromycin or ceftriaxone plus levofloxacin were not antagonistic. Furthermore, levofloxacin did not show synergy with azithromycin (or any other antimicrobial tested), indicating that the occasionally combined use of azithromycin plus levofloxacin for severe Legionella pneumonia is unlikely to provide benefit.
|
We have been applying synergy analysis for identification of combinations of existing antibiotics that are active against carbapanem-resistant Enterobacteriaceae (CRE). Interestingly, a large number of antibiotics are active against CRE when combined with colistin, including Gram-positive agents. The idea behind the latter finding is that colistin permeabilizes the Gram-negative outer membrane allowing prototypical Gram-positive agents such as linezolid, fusidic acid, and azithromycin in to access their targets in the bacterial cytoplasm. Importantly, in a large survey of CRE strains, we found that for 90% of strains there was at least one combination with what we call clinically relevant synergy, meaning that the minimal inhibitor concentrations for both antibiotics when used in combination was pushed into the susceptible range. These combinations will have to be validated in further pre-clinical and clinical studies; however, they offer hope of additional treatments for potentially otherwise untreatable pathogens.
The often cited emblem for a post-antibiotic world is the "Nevada strain." Isolated in 2017, this bacterial pathogen was resistant to all 26 available antibiotics tested by the CDC and therefore was considered pandrug-resistant. Unfortunately, the patient's infection was therefore untreatable at the time. The strain carries some highly troubling resistance elements. It is a CRE Klebsiella pneumoniae that expresses the NDM-1 metallo-carbapenemase that confers resistance to the broad range of beta-lactam antibiotics including the new drug, ceftazidime-avibactam. It also expresses a ribosomal methylase, making the strain impervious even to the newly approved aminoglycoside, plazomicin. We therefore applied high throughput synergy testing using D300 inkjet printing methodology to identify potential new treatment options.
In doing so, we identified multiple antibiotic combinations with activity against the Nevada strain. These included the combination of a number of drugs with colistin. The combination of ceftazidime-avibactam with aztreonam was found also to be highly synergistic, highlighting the potential use of this combination against even the most resistant NDM-1 strains. Demonstrating the power of synergy, none of the same drugs was active when tested alone. In addition, we found that apramycin and spectinomycin were highly active by themselves with compellingly low minimal inhibitory concentration values. The data suggested that these agents could be repurposed to treat highly drug-resistant infections and provide additional motivation for modification of the underlying molecular scaffolds to enhance activity further. You can read the full text of the final accepted article at the following link: "Synergistic Combinations and Repurposed Antibiotics Active against the Pandrug-Resistant Klebsiella pneumoniae Nevada Strain"
In doing so, we identified multiple antibiotic combinations with activity against the Nevada strain. These included the combination of a number of drugs with colistin. The combination of ceftazidime-avibactam with aztreonam was found also to be highly synergistic, highlighting the potential use of this combination against even the most resistant NDM-1 strains. Demonstrating the power of synergy, none of the same drugs was active when tested alone. In addition, we found that apramycin and spectinomycin were highly active by themselves with compellingly low minimal inhibitory concentration values. The data suggested that these agents could be repurposed to treat highly drug-resistant infections and provide additional motivation for modification of the underlying molecular scaffolds to enhance activity further. You can read the full text of the final accepted article at the following link: "Synergistic Combinations and Repurposed Antibiotics Active against the Pandrug-Resistant Klebsiella pneumoniae Nevada Strain"
(6) Evicting multidrug resistance plasmids to restore antibiotic susceptibility
In Gram-negative pathogens, most antibiotic resistance is encoded on large, low copy plasmids. These plasmids and their replication machinery are fascinating. Plasmids are subcategorized by their "incompatibility" group which in turn are related to the type of replication machinery and its regulatory control. Low copy is really low copy, at one to five copies per cell. Large is upwards of 100-300 kb. To ensure that daughter cells each receive low copy plasmid during cell division, segregation machinery is also employed. Resolvases take care of multimers. Plasmid addition systems kill off any bacteria foolish enough to loose their plasmid. Finally conjugation machinery (our favorite type IV secretion systems) enable transfer of the plasmid from one bacteria to another across genus and species. A great way to spread the joy.
We considered it should be possible to block functions critical for plasmid maintenance, thereby expelling plasmids and restoring antibiotic susceptibility. To establish if this were true, we conducted a high throughput screen using approximately 50,000 known bioactives and biologically uncharacterized compounds. In doing so, we identified a number of potent hits with specific mechanism of action and ability to restore susceptibility antibiotic. Several compounds completely stopped plasmid replication and some eliminated all plasmid within the limit of detection of all of our assays (qPCR and even more sensitive bacterial plating assau selecting for a resistance marker on the plasmid). Compounds restored susceptibility to meropenen in a CRE strain driving meropenem MICs will into susceptible range!
One prediction that we had is that potent anti-plasmid agents would indirectly lead to death of the host cell. Specifically as nearly all large, low copy resistance plasmids encode one or several plasmid addiction systems, plasmid eviction should not only restore susceptibility but efficient plasmid loss should also result in death of host bacteria cells. In other words, the bacterial cells will be killed by the stable plasmid addiction toxins that remain after plasmid curing and rapid loss of the cotranscribed unstable plasmid addiction antitoxins. Such was the case. Our findings for the first part of this work are described in PNAS manuscript: "Discovery of small-molecule inhibitors of multidrug-resistance plasmid maintenance using a high-throughput screening approach"
Here we specifically examine the effects on the IncFIA replicon (that's an incompatibility group) commonly found in CTX-M plasmids in E. coli ST-131, a epidemiologically rapidly spreading invasive group of E. coli. We are very excited to be collaborating with the Manetsch Laboratory to develop analogues of potent hits using medicinal chemistry approaches and definition of mechanism of action. We are very interested in what our findings will tell us about resistance plasmid maintenance and biology.
We considered it should be possible to block functions critical for plasmid maintenance, thereby expelling plasmids and restoring antibiotic susceptibility. To establish if this were true, we conducted a high throughput screen using approximately 50,000 known bioactives and biologically uncharacterized compounds. In doing so, we identified a number of potent hits with specific mechanism of action and ability to restore susceptibility antibiotic. Several compounds completely stopped plasmid replication and some eliminated all plasmid within the limit of detection of all of our assays (qPCR and even more sensitive bacterial plating assau selecting for a resistance marker on the plasmid). Compounds restored susceptibility to meropenen in a CRE strain driving meropenem MICs will into susceptible range!
One prediction that we had is that potent anti-plasmid agents would indirectly lead to death of the host cell. Specifically as nearly all large, low copy resistance plasmids encode one or several plasmid addiction systems, plasmid eviction should not only restore susceptibility but efficient plasmid loss should also result in death of host bacteria cells. In other words, the bacterial cells will be killed by the stable plasmid addiction toxins that remain after plasmid curing and rapid loss of the cotranscribed unstable plasmid addiction antitoxins. Such was the case. Our findings for the first part of this work are described in PNAS manuscript: "Discovery of small-molecule inhibitors of multidrug-resistance plasmid maintenance using a high-throughput screening approach"
Here we specifically examine the effects on the IncFIA replicon (that's an incompatibility group) commonly found in CTX-M plasmids in E. coli ST-131, a epidemiologically rapidly spreading invasive group of E. coli. We are very excited to be collaborating with the Manetsch Laboratory to develop analogues of potent hits using medicinal chemistry approaches and definition of mechanism of action. We are very interested in what our findings will tell us about resistance plasmid maintenance and biology.
(7) Efflux Pumps
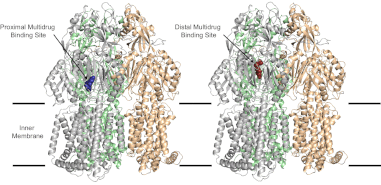
We collaborate with the structural biology laboratory of Ed Yu investigating multidrug efflux pumps in Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacterales. As part of this collaborative investigation, also in collaboration with NIH and Northeastern we found that the compound amotosalen, a psoralen compound used to inactivate pathogens in blood transfusion products is a facilely pumped out by pumps from all three species, in particular MexXY from Pseudomonas, AcrAB from E. coli and AdeABC from Acinetobacter..
Psoralens were found to be multidrug efflux substrates more generally, and were noted to follow eNTry rules for their respective gram-negative penetrance.
Psoralens were found to be multidrug efflux substrates more generally, and were noted to follow eNTry rules for their respective gram-negative penetrance.